1.1.1.189: prostaglandin-E2 9-reductase
This is an abbreviated version!
For detailed information about prostaglandin-E2 9-reductase, go to the full flat file.
Word Map on EC 1.1.1.189 
Reaction
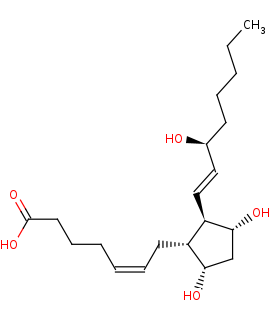 (5Z,13E)-(15S)-9alpha,11alpha,15-trihydroxyprosta-5,13-dienoate +
(5Z,13E)-(15S)-9alpha,11alpha,15-trihydroxyprosta-5,13-dienoate +
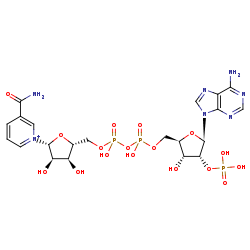 NADP+=
NADP+=
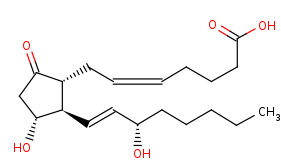 (5Z,13E)-(15S)-11alpha,15-dihydroxy-9-oxoprosta-5,13-dienoate +
(5Z,13E)-(15S)-11alpha,15-dihydroxy-9-oxoprosta-5,13-dienoate +
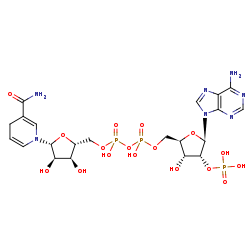 NADPH +
NADPH +
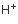 H+
H+
Synonyms
20-alpha-HSD, 20-alpha-hydroxysteroid dehydrogenase, 9-keto-prostaglandin E2 reductase, 9-ketoprostaglandin reductase, 9-KPR, CBR1, More, mPGES, PG 9-ketoreductase/carbonyl reductase, PGE 9-ketoreductase, PGE2 9-ketoreductase, PGE2-9-K, PGE2-9-ketoreductase, PGE2-9-OR, prostaglandin 9-ketoreductase, prostaglandin 9-ketoreductase/carbonyl reductase, prostaglandin E 9-ketoreductase, prostaglandin E2 9-ketoreductase, prostaglandin E2-9-oxoreductase, prostaglandin-E2 9-keto reductase, reductase, 15-hydroxy9-oxoprostaglandin
ECTree
Sequence
Sequence on EC 1.1.1.189 - prostaglandin-E2 9-reductase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
CBR1_BOVIN
277
0
30533
Swiss-Prot
other Location (Reliability: 5)
CBR1_HUMAN
277
0
30375
Swiss-Prot
other Location (Reliability: 4)
CBR1_MACFA
277
0
30473
Swiss-Prot
Mitochondrion (Reliability: 4)
CBR1_MOUSE
277
0
30641
Swiss-Prot
Mitochondrion (Reliability: 5)
CBR1_PIG
289
0
31693
Swiss-Prot
Mitochondrion (Reliability: 3)
CBR1_PONAB
277
0
30391
Swiss-Prot
other Location (Reliability: 5)
CBR1_RABIT
277
0
30452
Swiss-Prot
Mitochondrion (Reliability: 4)
CBR1_RAT
277
0
30578
Swiss-Prot
other Location (Reliability: 4)
PE2R_RABIT
323
0
36670
Swiss-Prot
other Location (Reliability: 2)
Q9TSA9_PIG
65
0
6923
TrEMBL
other Location (Reliability: 1)
A0A5B7BAK0_DAVIN
281
0
30920
TrEMBL
Mitochondrion (Reliability: 4)
A0A0C1C1D0_9BACT
241
0
26248
TrEMBL
-
A0A0B2RJ58_GLYSO
303
0
33173
TrEMBL
other Location (Reliability: 3)
F8L0N3_PARAV
Parachlamydia acanthamoebae (strain UV7)
231
0
24993
TrEMBL
-
Q09E76_STIAD
Stigmatella aurantiaca (strain DW4/3-1)
234
0
26134
TrEMBL
-
A0A2N5WD83_LACLL
261
0
28700
TrEMBL
-
Q097P7_STIAD
Stigmatella aurantiaca (strain DW4/3-1)
234
0
25733
TrEMBL
-
A0A0B8NGM8_9NOCA
281
0
29530
TrEMBL
-
A0A084FV16_PSEDA
325
0
36940
TrEMBL
other Location (Reliability: 3)
A0A6J8BIX3_MYTCO
281
0
30784
TrEMBL
Mitochondrion (Reliability: 3)
B7P728_IXOSC
73
0
8188
TrEMBL
other Location (Reliability: 2)
B9SFU9_RICCO
296
0
32892
TrEMBL
other Location (Reliability: 2)
B6R2U0_9HYPH
219
0
23470
TrEMBL
-
A0A1Z5K650_FISSO
276
0
30071
TrEMBL
other Location (Reliability: 4)
A0A1Z5JLW5_FISSO
276
0
30188
TrEMBL
other Location (Reliability: 3)
B9RDN5_RICCO
544
0
59210
TrEMBL
other Location (Reliability: 5)
A0A0B2RU65_GLYSO
77
0
8367
TrEMBL
other Location (Reliability: 4)
B9RDN3_RICCO
333
0
36598
TrEMBL
Secretory Pathway (Reliability: 4)
A0A812B6N7_SEPPH
774
0
84138
TrEMBL
Mitochondrion (Reliability: 5)
A0A812E5X8_SEPPH
238
0
26088
TrEMBL
other Location (Reliability: 3)
html completed




 results (
results ( results (
results ( top
top









